An overview of methods used in zebrafish research.
Zebrafish (Danio rerio) is a fresh water fish that inhabits rivers in India, Pakistan and other places in Asia. In the past two decades, Zebrafish has become one of the preferred in vivo model organisms for studying diverse processes like regeneration [8], embryogenesis [9, 10], autophagy [11], behaviour [12, 13] and sleep. Recently, Zebrafish has sparked interest in other branches of the biomedical research because of its emerging potential for drug discovery in various disease models like cardiotoxicity [14] and dyslipidemia and atherosclerosis [15]. For instance, zebrafish with the mutated MYO18B gene can serve as a model for human myopathy [16]. Metzner A et al screened compound libraries on zebrafish pkd2 mutant embryos to identify therapeutic candidates for autosomal dominant polycystic kidney disease [17]. This review addresses a selection of the most commonly used methods in zebrafish research. While the general breeding and handling of zebrafish are detailed in the "Zebrafish Book" [18], this article highlights the important tools and methodology available in zebrafish research such as microinjection, morpholino knockdown and transplantation, and discusses their applications.
Zebrafish has become a favorite model of choice in the biomedical research covering a broad array of topics ranging from developmental biology and morphogenesis to neurosciences, regeneration and aging in the past decades. The ease of maintenance, the large number of offsprings, an option to conduct genetic screens, and the optical clarity of developing embryos constitute just a few factors making zebrafish an attractive model organism to work with. While the main advantage in working with zebrafish is the possibility to conduct high resolution in vivo imaging, the community was lagging behind the other model organisms such as the fruit fly (Drosophila melanogaster) and the mouse because of a lack of appropriate genetic tools for generating transgenic animals and the efficient gene knock-outs. However, in the past few years, these gaps have been closing due to the rapid advance in technology. New emerging tools such as CRISPR, base or prime editing, and zinc finger nucleases allowing efficient gene knock-out or MAZe and brainbow transgenic technologies, make it possible to assess the complexity of the developmental processes and gain insights into the mechanisms of diseases in a vertebrate. Excellent protocols and reviews exist about CRISPR application in zebrafish research [19-21] and the technology is evolving rapidly; thus it is not discussed here.
In addition, human and zebrafish genomes share about 70% homology, suggesting the conservation among a large number of genes and genetic pathways. Because of the availability of technological platforms, zebrafish is being used in a medium-throughput drug screening in the context of the whole organism (as compared to cell-free or single cell systems). The present review focuses on the pertinent methods, starting from injections, immunostaining and in situ labeling and then discussing the gene expression knock-down, targeted mutagenesis, transgenic tools and mosaic experiments.
More recently, it has been suggested that the commonly used anaesthetic MS-222 should be avoided since zebrafish find it distressing [22, 23], although it is still being used [11].
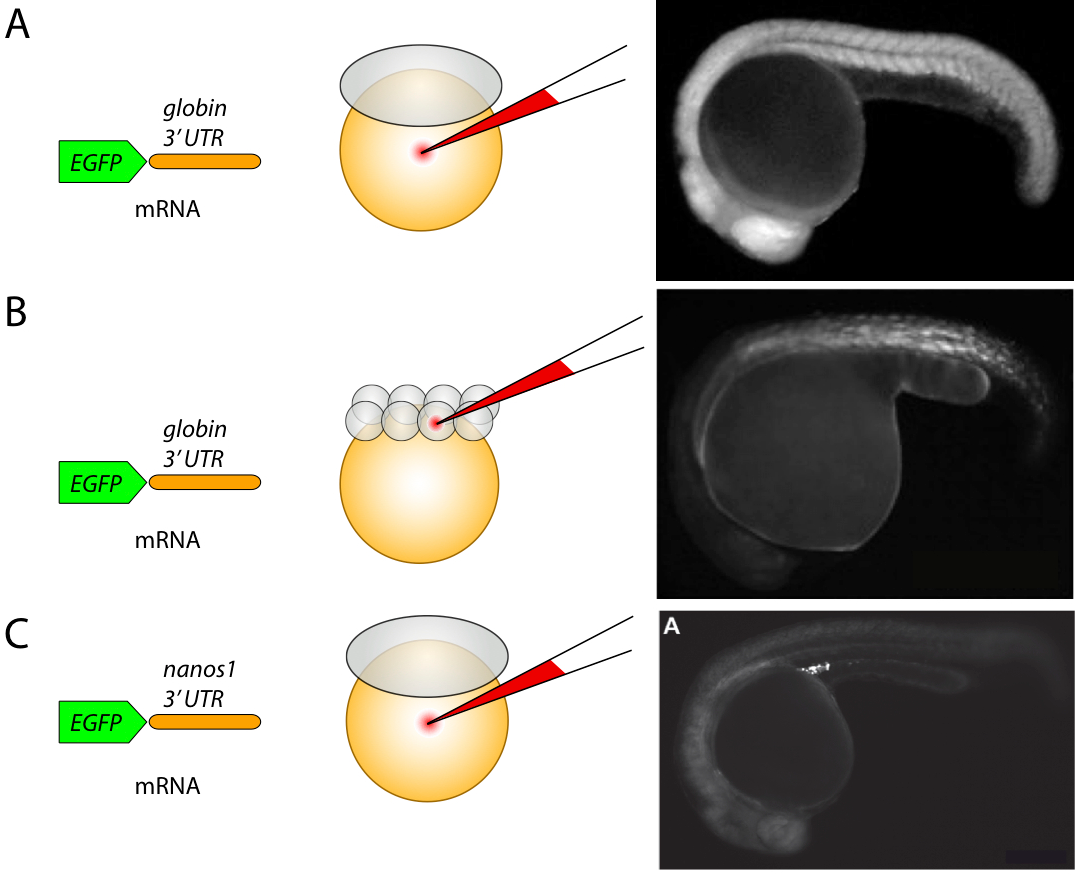
Injection is the most frequently used technique when working with zebrafish. During injection, a certain substance such as the nucleic acid, protein, or a certain drug is introduced into the embryo, usually at the early stages of development (Figure 1). Injections are done with glass needles filled with the material to be delivered. Micromanipulators normally control the needles and can change the position of a needle relative to the embryo with ease. Air pressure provided by an external device is used to release the content into the embryo. Injection allows rapid delivery of the material into embryos. An experienced researcher can inject a few hundreds of embryos in one hour. Detailed injection protocol is available in [24] and its movies can be viewed at [25].
Injections are most frequently used for transient overexpression of proteins in the embryos. Transient overexpression during early zebrafish development (up to 3 days) is achieved by introducing the mRNA encoding the protein of interest into the embryos during the first two hours of development. To stabilize the injected mRNA transcript, the regulatory mRNA regions, such as 3' untranslated regions (UTR) are used (Figure 1). These include the Poly A sequences such as those present in the beta-globin gene from Xenopus laevis, which stabilize the mRNA transcript in every cell of the embryo containing this mRNA (Figure 1A and B). For certain cell types, such as primordial germ cells, specific UTRs such as those found in the 3'-UTR of the Nanos1 gene can be used to stabilize the mRNA and to promote the efficient protein translation specifically in these cells while degrading it in the somatic cells [26] (Figure 1C). Another example for the 3'-UTR known to play a role in mRNA localization is found in squint mRNA, which is confined to the dorsal blastomeres as early as at the four cell stage [27].
To drive the protein expression in every cell of the developing embryo, the mRNA corresponding to a particular protein is injected into the yolk of the one-cell stage embryo (Figure 1A). For example, Zhao N et al injected FB-GFP and N × HA-mCh-H2B mRNAs into the yolk of 1-cell stage embryos and tracked the fluorescence in 4D [28]. To generate mosaic embryos, in which the protein is expressed in a subpopulation of cells, the mRNA is injected into a single blastomere of the 8-cell stage embryo or at the later stages (Figure 1B). Another strategy used to achieve mosaic protein expression is by injecting the DNA plasmids into a single blastomere of the one-cell stage embryo or later on (directly into a specific cell), however this approach is less efficient and takes longer for protein synthesis as compared to the mRNA injection.
While transient expression of proteins via mRNA injection is a quick and easy way to study protein localization and function in zebrafish at the early stages of development, this approach is not suitable when the later developmental stages are of interest or when a protein must be expressed in a particular tissue or a cell type.
Cancer cells can also be injected zebrafish embryos to generate xenograft models. For example, Lundby A et al injected A549 cells into zebrafish embryos to study the regulation of invasiveness of A549 cells by the EGF receptor and its mutant [29].
In situ hybridization technique is one of the oldest methods used in zebrafish research. It enables the investigation of gene expression patterns in intact embryos or in sections [30]. During the in situ hybridization procedure, an antisense mRNA probe is designed to recognize and bind the endogenous transcript, which is later detected by the color-based or fluorescence-based assay. The classical approach utilizes a color-based labeling procedure and the signal is visualized with a light microscope (Figure 2A). Recently fluorescent-based detection techniques had been optimized for the use in zebrafish, enabling the simultaneous detection of three probes with the regular approach, or up to five probes using the HCR amplification (Figure 2B and C) [4, 31, 32]. Fluorescent-based in situ hybridization is the preferred method of signal detection because of the higher sensitivity and the simultaneous detection of multiple probes, while the color-based detection is limited to two probes. However, the classical color staining does not require a confocal microscope setup and can be used when the confocal microscope is not available.

Immunostaining is a powerful method for detecting the presence and the localization of an endogenous protein. The main obstacle in performing successful immunolabeling is the antigen retrieval. Another difficulty arises from a relatively low number of the commercially available high quality antibodies developed for the use in zebrafish. A recent paper proposed an improved method for treating embryos for the efficient antigen retrieval (Figure 2D) [5].
A number of transgenic tools have been adapted from other organisms and optimized for the use in zebrafish allowing stable protein expression and the conditional targeting of proteins for expression in defined tissues.
The most popular system for generating transgenic animals in zebrafish is the Tol2 transposon system, for example [8], originated from the medaka fish [33, 34]. The construct for introducing the DNA into the zebrafish genome typically consists of two minimal cis regulatory sequences from the Tol2 element positioned at the 5' and the 3' of a minimal promoter followed by a fluorescent protein (Figure 3). The Tol2 containing vector is capable of accommodating the DNA inserts of up to 11 kb [33]. Such DNA construct is injected together with the mRNA encoding for a transposase gene directly into the cell of the one-cell stage embryos (Figure 3). These embryos are raised and the adult fish are screened to identify the founders, in which the insertions were created in the germ-line that can be transmitted to the next generation (depicted as F1 in Figure 3). For example, Wan Y et al generated transgenic line Tg(β-actin2:H2B-HaloTag) through the construct pDestTol2CG-β-actin2-H2B-HaloTag-pA [35]. Gu Q et al used a Tol2-based system to generate mutant zebrafish to investigate the fate determination of hematopoietic stem and progenitor cells [36].

Another strategy for the regulated protein expression that has been recently adapted for the use in zebrafish is the cre-mediated site-specific recombination [1]. Cre-mediated recombination allows the induction of protein expression in a temporally controlled fashion (Figure 4A). Cre-recombinase expression can be further regulated by the heat-shock promoter allowing the high temporal and spatial precision of this protein expression. Such a strategy has been utilized in the MAZe transgenic system for spatial-temporal protein expression in the mosaic embryos [37] (Figure 4B). The MAZe system combines the use of Cre-recombinase expressed under the heat-shock promoter and the GAL4/UAS system adapted for the use in zebrafish. Following the heat shock induction, Cre-recombinase is produced, resulting in the excision of the cassette enclosed by the lox P elements, thereby allowing the synthesis of the Gal4VP16 protein that in turn binds to the UAS promoter driving the expression of the specific protein. This method is particularly useful when inducing the expression of dominant-negative proteins in a selected population of cells at the defined time point.

Recent studies have used direct delivery of Cas9 into zebrafish embryos at the earliest stage of development in order to induce double stranded breaks [38] or prime editing [39].
Production of specific mutations is mediated via the non-homologous end joining (NHEJ), which is applied in zebrafish in order to knock-in a selected DNA sequence at a specific genomic region with CRISPR/Cas9 [40]. In particular, the selected sequence is co-delivered together with a CRISPR/Cas9 construct. Moro A et al injected directly Cas9 mRNA and gRNAs to generate mutants at the ctgfa miRNA responsive elements [41]. Gu Q et al used a CRISPR/Cas9-based system to knockout apoa1bp2 and srebf2 genes in zebrafish to investigate the fate determination of hematopoietic stem and progenitor cells [36]. There are several different homology-based approaches to incorporating exogenous DNA together with the selected sequences of interest. One of them, microhomology-based repair (mHDR) knock-in method applies CRISPR/Cas9 to insert short single-stranded sequences [42]. Larger sequences, such as constructs encoding fluorescent proteins, have also been incorporated with short homology sequences. Short homology elements have been used to fuse green fluorescent protein with the sequence encoding Keratine type 1 [43]. Furthermore, a new variation of mHDR, called seamless integration strategy, has been lately introduced for incorporating mCherry [44]. Another method, large homology-directed repair (lHDR), is based on the insertion of large DNA sequences with CRISPR/Cas9 for making knock-in models in zebrafish. For instance, a large albino gene was inserted to induce pigmentation in albino animals [45]. Several techniques for improvement of HDR techniques have recently been suggested. For instance, the conformation of donor DNA sequences applies a circular plasmid [46] or restriction endonucleases [40]. Another method, which enhances HDR, is characterized by the inhibition of the main NHEJ components [47]. Alternatively, the introduction of cas9 mRNA instead of Cas9 protein has been shown to improve the generation of knock-in zebrafish models [48].
K Petri et al injected a prime editing guide RNA and a Cas9 nickase fused to an engineered M-MLV reverse transcriptase into zebrafish embryos to introduce somatic mutations with frequencies as high as 30% [39].
The standard approach to block specifically the function of a gene in a zebrafish embryo at the early stages of development is with the modified antisense oligonucleotides called morpholinos [49, 50]. Morpholinos are synthetic molecules, about 25 nucleotide length, and are available at Gene Tools [51]. Two types of morpholinos are commonly used to interfere with the protein expression: the ATG morpholinos and the splice morpholinos. The ATG morpholinos act by blocking the initiation of protein translation at the ribosomes, thereby rendering the embryos devoid of a particular protein [52]. Figure 5A shows an example of a three-day old embryo missing pectoral fins as the result of the morpholino treatment against the fgf24 gene [6]. Splice morpholinos, for example, morpholinos targeting gpr124 (ACTGATATTGATTTAACTCACCACA) [53], bind and interfere with the RNA splicing procedure, resulting in the truncated form of a protein. Therefore, splice morpholinos could be as well used to study the function of a particular protein domain. For example, Wan Y et al injected antisense morpholino oligonucleotides from Gene Tools to knock-down a specific gene [35]. Gu Q et al used morpholino antisense oligos against apoa1bp2 and srebf2 in zebrafish to investigate the fate determination of hematopoietic stem and progenitor cells [36].

Target protectors morpholinos have been recently developed to interfere with the function of the endogenous microRNAs (miRNA) specifically at their targets. These morpholinos are complimentary to the miRNA binding sequences present in the target mRNAs and act by preventing the miRNA binding to its target, therefore stabilizing the mRNA transcript [54]. Similarly, morpholinos can be used to inhibit the miRNA function directly [55]. The general considerations in handling morpholinos and the interpretation of the experimental results are described in detail in [52, 56].
Morpholinos represent a highly efficient strategy to interfere with the gene or miRNA function with high precision, however there are several pitfalls concerning morpholinos that must be taken into consideration. Recently, it has been suggested that the effects of morpholinos are not specific and up to 80% observed Morpholino-induced phenotypes are due to false-positives [57].
- Morpholinos are normally injected into the yolk and therefore could equally affect every cell in the developing embryo and thereby might compromise the proper development in cases when the function for the disrupted protein is required ubiquitously. This problem however, could be avoided by injecting the morpholino into a single blastomere (Figure 1B) in an attempt to target the cells of interest more precisely.
- The effect of morpholinos lasts only the first few days of development (up to 5 days [52] ), making it unsuitable for studying the function of a gene at later stages of development.
- Morpholinos are not effective when the cognate maternal protein is present.
- While a morpholino eliminates protein function, the corresponding mRNA remains present in the embryo. In cases when the mRNA plays another role in addition to serving as a template for protein translation, it could potentially lead to the non-specific effects, especially if the levels of mRNA are important.
While until recently there was no reliable method of inducing targeted mutagenesis in zebrafish, two papers published in 2008 adapted the use of zinc finger nucleases (ZFN) for creating the targeted double-stranded breaks in the zebrafish genome. In two independent studies, the authors have demonstrated the use of ZFN to generate the breaks, which were then repaired by the non-homologous end joining, resulting in small insertions and deletions. ZFN represents a chimeric fusion protein consisting of a zinc finger protein (ZFP) and the cleavage domain from the FokI endonuclease [58]. The DNA binding specificity is defined by the ZFP, which can be engineered to recognize a variety of the target DNA sequences [58] (Figure 5B and [59, 60] ). The important part in designing ZFN is the ZFP optimization for the target recognition. Detailed protocols and the additional background are available at ZFIN site [61] and the advice on how to use the OPEN platform for engineering a zinc finger array is given in [62]. Additional information on how to locate ZFN target sites can be found at [63].

Transplantation is one of the oldest techniques employed in developmental biology. During transplantation, a group of cells is removed from one embryo, called the donor embryo and introduced into another embryo, which is called the acceptor embryo (Figure 6A). Transplantation technique is essential in addressing some of the important questions in developmental biology, such as whether a gene function is cell autonomous or not. In the experiments studying the gene function, the mutant cells are transplanted into a wild type embryo and conversely, the wild type cells are transplanted into a mutant embryo and the phenotypes of the transplanted cells and those of the host embryo are compared. Transplantation can also be used when the morpholino injection is lethal. In that case, a group of cells from the embryo injected with a morpholino are transplanted at the early stages of development into the wild type embryo, thereby addressing the effect of the protein knockdown on the cells in a wild type environment. Transplantations are also used to study the effect of a diffusible factor, such as a growth factor or a chemokine on neighboring cells. An example of how transplantations are used to address the mechanisms underlying the neuron-glia interactions is presented in [7, 64] (Figure 6B).
The title of this article was changed from "Current Methods in Zebrafish Research" to "Zebrafish Research Methods" in October 2019.
- Ellingsen S, Laplante M, König M, Kikuta H, Furmanek T, Hoivik E, et al. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799-811 pubmed
- ZFIN ID: ZDB-IMAGE-070503-669 http://zfin.org.
- Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann C, Voz M, et al. Reciprocal endoderm-mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011-21 pubmed
- The Zebrafish Book. Available from: zfin.org/zf_info/zfbook/cont.html
- Microinjection of Zebrafish Embryos to Analyze Gene Function. Available from: www.jove.com/details.php?id=1115
- Köprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877-85 pubmed
- Gore A, Maegawa S, Cheong A, Gilligan P, Weinberg E, Sampath K. The zebrafish dorsal axis is apparent at the four-cell stage. Nature. 2005;438:1030-5 pubmed
- Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 1994;10:73-4 pubmed
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8 Suppl 1:S7 pubmed
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141-58 pubmed
- Nasevicius A, Ekker S. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216-20 pubmed
- Gene Tools LLC. Available from: www.gene-tools.com/
- Choi W, Giraldez A, Schier A. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271-4 pubmed
- Flynt A, Li N, Thatcher E, Solnica Krezel L, Patton J. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259-63 pubmed
- Porteus M, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967-73 pubmed
- Zinc Finger Consortium. Available from: zincfingers.org/default2.htm
- Tissue Targeted Embryonic Chimeras: Zebrafish Gastrula Cell Transplantation. Available from: www.jove.com/details.php?id=1422
- Materials and Methods [ISSN : 2329-5139] is a unique online journal with regularly updated review articles on laboratory materials and methods. If you are interested in contributing a manuscript or suggesting a topic, please leave us feedback.
- method
- Base Editing
- Behavioral Phenotyping in Rats and Mice
- CRISPR and Genomic Engineering
- Cloning and Expression Vectors, and cDNA and microRNA Clones Companies
- Current Approaches in C. elegans Research
- Huntington's Disease Animal Models
- Inflammasome
- Inteins
- Laboratory Mice and Rats
- Optogenetics
- Stem Cell Research Using Mouse Models
- Xenopus laevis as a Model System
