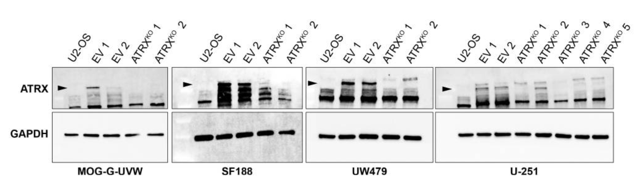
This is a knockout-examined antibody summary, based on the publication "ATRX loss induces multiple hallmarks of the alternative lengthening of telomeres (ALT) phenotype in human glioma cell lines in a cell line-specific manner", as cited below [1]. This article indicates that this antibody recognizes ATRX and also stain other bands in all cell types tested in western blot. Labome curates formal publications to compile a list of antibodies with unambiguous specificity within Validated Antibody Database (VAD).
Cell Signaling Technology (CST) re-tested this antibody after Labome communicated the finding from this article to CST. CST examined this antibody on lysates from six cell lines: U-2 OS, SAOS-2, SF295, LN-18, MIA PaCa-2, and SH-SY5Y with a 3-8% Tris-Acetate gel. This antibody recognized a prominent band with the approximate molecular weight from SF295, LN-18, MIA PaCa-2, and SH-SY5Y cell lines. The western blot background was normal, not as prominent as in Figure 1. Thus the antibody is specific. For further information, please contact CST directly.
Rabbit monoclonal IgG
Company: Cell Signaling
Antibody: ATRX
Catalog number: 14820
Summary: Rabbit monoclonal IgG against a synthetic peptide corresponding to residues surrounding Leu1189 of human ATRX protein. Reacts with human by western blot, immunoprecipitation and Chromatin IP.
Western blot. The antibody recongizes the correct band and also shows other nonspecific bands in all cell types tested.
Control and ATRX-KO MOG-G-UVW, U-251, SF188, and UW479 cell lines. Cells were lysed in RIPA buffer (Cell Signaling, Cat#9806) containing a cocktail of protease inhibitors (Roche).
5% milk in TBST.
1:1,000 dilution in 5% milk in TBST overnight.
Anti-rabbit HRP (Cell Signaling, 7074) for one hour.
Signal was detected using Clarity ECL (BioRad). Blots were either exposed to film or developed using a ChemiDoc Touch (BioRad).
- If you are aware of any publication with knockout studies validating a monoclonal or recombinant antibody, either purchased from a supplier or developed by the author(s), please notify us through feedback.
