A compilation of T cell surface markers and B cell surface markers at different stages of development and a summary of high-quality antibodies against these markers cited among the over 60,000 formal publications in Labome's Validated Antibody Database.
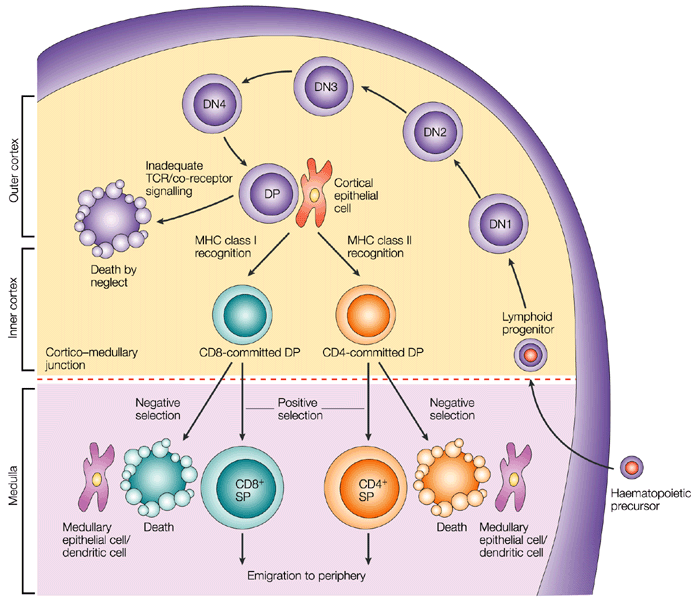
It is well established that CD4- CD8- T cell precursors migrate to the thymus where they undergo the following phenotypical stages: CD44+ CD25- (DN1), CD44+ CD25+ (DN2), CD44- CD25+ (DN3), and CD44- CD25- (DN4), followed by the progression of DN4 cells into the double-positive CD4+ CD8+ T cells [5] (Figure 1). Infection by various pathogens causes activation and proliferation of naïve T cells, which differentiate into lineages with effector and memory fates. Naive CD4+ T cells recognize antigens presented by major histocompatibility complex (MHC) class II on antigen-presenting cells. Depending on the specific stimuli, the CD4+ T cells can differentiate into various subtypes, including the helper TH1, TH2 and TH17 cells and regulatory T cells (Tregs). A subset of TH2 cells differentiate into allergic disease-related TH2A cells, with a CD45RBlow CD27− phenotype and coexpression of the chemoattractant receptor CRTH2, the natural killer cell marker CD161, and the homing receptor CD49d [6]. Memory T cells vary in their surface receptor expression, effector and trafficking abilities. There are four major subsets of memory T cells: central memory, effector memory, tissue-resident memory and stem memory T cells. Multiple signals regulate the differentiation of CD4+ T cells into central and peripheral memory cells. CD4+ T central memory cells express CD62L and CCR7, which are important for their migration [7]. The peripheral T stem cell memory cells express CXCR3 and CD95 molecules. In addition, both naive and memory T-cell subsets express a variety of functional molecules (Table 1).
| Process | Antigen | Function |
|---|---|---|
| Costimulation/Survival | CD27 | Costimulation |
| CD28 | Costimulation | |
| CD127 | IL-7 signaling | |
| PD-1 | Inhibition of effector function | |
| CD122 | IL-2/IL-15 signaling | |
| CD132 | γc cytokine signaling | |
| KLRG-1 | Inhibition of effector function | |
| Activation | HLA | DR Peptide presentation |
| CD38 | Calcium flux/signal transduction | |
| CD69 | Proliferation | |
| Adhesion | CD11a | Adhesion to APC/endothelium |
| CD58 | Adhesion to APC | |
| CD99 | Transendothelial migration | |
| Migration | CD62L | Secondary lymphoid tissues homing |
| CD103 | Gut homing | |
| CCR4 | Chemokine response/TH2 associated | |
| CCR5 | Homing to inflamed tissues | |
| CCR6 | Chemokine response/TH17 associated | |
| CCR9 | Gut homing | |
| CCR10 | Skin homing | |
| CXCR3 | Homing to inflamed tissues | |
| CXCR4 | Homing to bone marrow | |
| CLA | Skin homing | |
| Cytolytic molecules | Granzyme A | Cleavage of cellular proteins |
| Granzyme B | Cleavage of cellular proteins | |
| Perforin | Pore-forming | |
| Miscellaneous | CD161 | Regulation of proliferation/cytotoxicity |
| IL-18Ra | 18Ra Response to IL-18 | |
| c-Kit | Response to SCF | |
| CD130 | Response to IL-6 |
The second major group of T cells, CD8+ T cells, mediates direct killing of antigen-presenting target cells. Naive CD8+ T cells are activated upon recognition of antigens presented by MHC class I on dendritic cells in the spleen or lymph nodes. Activated CD8+ T cells expand and become effector CD8+ T cells. CD8+ T cells tend to be evaluated during the study for tumor-infiltrating T cells. For example, Vodnala SK et al. evaluated the effect of overabundance of potassium in the tumor microenvironment on CD8+ T cell stemness and dysfunction in tumors [9].
The majority of the T cells bear α and β chains in their T cell receptor (TCR). However, there is a population of T cells, which have TCR formed by γ and δ chains. These cells, gamma delta T cells, bind to BTN2A1 and BTN3A1 [10], are significantly enriched in epithelia [11, 12]. Gamma delta T cells regulate immune responses by various mechanisms, including suppression of effector T cell and TH1 cell functions, blockage of neutrophil influx and regulation of antigen-presenting cell activity.
Naive T cells are considered as precursors of the majority of antigen-activated T cell subpopulations. Human naïve CD4+ T cells express CD45RA, CCR7, CD62L and CD27. Upon recognition of antigens presented by major histocompatibility complex (MHC) class II on antigen-presenting cells, naïve CD4+ T cells undergo proliferation and differentiation into functionally different T cell subsets including IFN-γ producing helper T cell-1 cells (TH1), IL-4-producing TH2, IL-17-producing TH17 cells, and inducible regulatory T cells (iTregs) (Figure 2). Each T cell subset expresses specific transcription factors, such as T-bet (TH1), GATA3 (TH2), RORγt (TH17), and Foxp3 (CD25+ Tregs) [13]. Grandclaudon M et al used IL-12Rb2 as a TH1 cell marker [14]. With regard to TH17 cells, their differentiation is under control TGF-β and IL-6-induced differentiation, IL-21-induced activation, and IL-23-regulated stabilization [15, 16]. As to iTregs, FOXP3 was found to an important marker of natural CD4+ CD25+ regulatory T cells. Moreover, transfection of CD4+ CD25- T cells with Foxp3 stimulates their regulatory activity [17]. In addition, TGF-β was found to be crucial for the differentiation of naive CD4+ T cells into Foxp3+ Tregs [18]. Also, IL-2 is commonly required for TGF-β-regulated iTreg differentiation [19]. D Mathew et al examined the six CD4+ T cell subtypes in Covid-19 patient blood: naïve (CD45RA+ CD27+ CCR7+ CD95-), central memory (CD45RA- CD27+ CCR7+), effector memory 1 (CD45RA- CD27+ CCR7-), effector memory 2 (CD45RA- CD27- CCR7+), effector memory 3 (CD45RA- CD27- CCR7-), and EMRA (CD45RA+CD27- CCR7-) in addition to circulating CD4+ T cells (CD45RA- PD1+ CXCR5+) and activated circulating CD4+ T cells (CD38+ICOS+) [20].

With regard to cytotoxic T cells, there are several peripheral subsets of different subsets of CD8+ T cells based on the expression of CD45RA and CCR7: a CD45RA+ CCR7+ subset of naive cells, a CD45RA- CCR7+ subset of antigen-experienced memory T cells, a CD45RA- CCR7- effector memory cell subset, and a CD45RA+ CCR7− subset of differentiated, antigen-experienced effector cells. Also, there are effector memory CD8+ T cells expressing CD69 and CD103 and residing in non-lymphoid tissues [21, 22]. A subpopulation of CD8+ T cells shows a memory cell phenotype: CD62L- /+ CCR7+ CD27- /+ . Activated cytotoxic CD8+ T cells downregulate expression of L-selectin and CCR7 and upregulate surface expression of CD44, LFA-1 and/or α4β1 integrin. In addition, there are CD8- cytotoxic T cells: CD4+ cytotoxic T cells and gamma delta T cells [23]. Hashimoto K et al shows that supercentenarians have much higher level of CD4+ cytotoxic T cells in blood circulation than young people [23]. D Mathew et al examined the six CD8+ T cell subtypes in Covid-19 patient blood with CD45RA, CD27, CCR7, and CD95 cell surface markers: naïve (CD45RA+ CD27+ CCR7+ CD95-), central memory (CD45RA- CD27+ CCR7+), effector memory 1 (CD45RA- CD27+ CCR7-), effector memory 2 (CD45RA- CD27- CCR7+), effector memory 3 (CD45RA- CD27- CCR7-), and EMRA (CD45RA+CD27- CCR7-) [20].
There are several additional phenotypical markers expressed in both human and mouse Tregs. They include CTLA-4, CD103, GITR and OX40. In particular, CTLA-4 is important for both inhibitory functions and homeostasis of Tregs. Intracellular expression of CTLA-4 was observed in CD4+ CD25+ human Tregs [24]. Another marker, integrin α (CD103) is expressed by Tregs and CD4+ CD25+ CD103+ Tregs were demonstrated to produce IL-10 actively [25]. In addition, GITR (CD357) is expressed in CD4+ CD25+ human Tregs in peripheral blood [26]. Also, OX40 (CD134) was shown to stimulate the proliferation of CD4+ FoxP3+ Tregs [27]. Moreover, OX40 stimulates migration of Tregs into the peripheral lymphoid and other tissues during inflammation [28].
Several specific T cell subsets, including follicular B helper T cells (TFH), follicular regulatory T cells (TFR) and cytotoxic CD8+ T cells, reside in the germinal centers and regulate the B cell proliferation [29]. Among these T cell subpopulations, TFH cells belong to CD4+ T cells and assist follicular B cells located in secondary lymphoid tissues, such as lymph nodes, spleen, and tonsils. Concerning the specific markers, high expression of CXC-chemokine receptor 5 (CXCR5) characterizes TFH cells [30]. Its interaction with CXC-chemokine ligand 13 (CXCL13) produced by follicular stromal cells mediates the homing of TFH cells into lymphoid follicles [30]. The development of TFH cells is strongly dependent on IL-2 production, as naїve IL2-secreting CD4+ T cells are destined to differentiate into TFH cells, while other CD4+ T cells, which do not produce IL-2, develop into non-TFH cells [31]. In addition to a universal T cell marker Thy1 (CD90) and CXCR5, TFH cells express ICOS and PD-1 molecules. Upregulation of CXCR5 expression stimulates TFH cells to migrate into the germinal centers, where these cells stabilize their phenotype by contacts with local B cells via ICOS-ICOSL binding [32]. Concerning the regulation of TFH functions, γδ T cells (TCRγδ+ CXCR5+ T cells), which also reside in the lymph nodes, have recently been shown to present antigens to TFH cells and induce their activation [33].

Cellular interactions with TFH cells regulate the proliferation and maturation of B cells in the germinal centers [34]. Besides, TFH cells secrete Il-4 and IL-21 cytokines, which are crucial for the functioning of the germinal centers [34]. Moreover, in the germinal centers, TFH cells are represented by at least two distinct subpopulations: IL-21+ T cells regulating the selection of high-affinity B cells and IL-4+ T cells promoting differentiation of plasmocytes [35]. A third type, secreting IL-4, IL-5, and IL-13, directs the class switching of B cells from IgG1 to high-affinity IgE during anaphylaxis, and coexpress transcription factors BCL6 and GATA3 [36]. Several studies have shown that chronic viral infection strongly induces differentiation of TFH, which leads to non-specific B cell activation [37].
In addition to TFH cells, researchers have identified TFR cells in the germinal centers. This subset of T cells expresses Foxp3 and also regulates the activity of germinal centers [38]. TFR cells suppress the proliferation of B cells and the production of IgM and IgG antibodies [38, 39] and diminish the secretion of IL-4 and IL-21 by TFH cells in the germinal centers [40].
The standard methods for measurement of T cell immune responses include Enzyme-Linked Immuno Spot assay (ELISpot), Intracellular Cytokine Staining assay (ICS), Tetramer assay and Flow Cytometry. The ELISpot and ICS assays apply in vitro stimulation to analyze the cytokine expression profiles of responding cells. The ELISpot method detects spots of cytokines secreted by individual cells, and ICS examines surface markers and produced cytokines. Multiple approaches can measure the proliferation of T cells in response to specific antigens, including thymidine incorporation assay, flow cytometric analysis of CD38 expression or ELISA detection of BrdU incorporation into DNA of proliferating T cells.
Many solid tumors are infiltrated by T cells such as CD8+ cells. Cytotoxic CD8+ T lymphocytes recognize an antigen-MHC complex on tumor cells, get activated and release perforin and granzymes causing apoptosis in target tumor cells [41]. Tumor antigen-specific T cells are used for the development of new immunotherapeutic methods. Cytotoxic T cells expressing chimeric antigen receptors (CAR T cells) have been effectively used for hematological malignancies [42]. In addition to CAR T lymphocytes, bispecific T cell engagers (BiTEs), represented by two scFvs, bind to both CD3 on T cells and a tumor cell antigen. One of the BiTEs, blinatumomab, was approved to treat B cell leukemia [43].
| T cellular subsets | Markers | Functions | References |
|---|---|---|---|
| Cytotoxic T cells | CD8+ | Exhibit cytotoxicity against tumor cells. | [44] |
| CD4+ TH9 T helpers | CD4+ | Secrete IL-9, IL-10 and IFNg and inhibit tumor growth. | [45] |
| CD4+ Tregs | CTLA-4+ GITR+ PD-1+ CCR+ CCR4+ CXCR4+ GITR+ LAG3+ OX40+ ICOS+ | Suppress anti-tumor immune response. | [46-48] |
| CD8+ Mucosal-associated invariant T cells | Rearranged TCRβ chains with Vβ gene segments | Production of TH1 -mediated cytokines and direct lysis of neoplastic cells. | [49, 50] |
| CD8+ memory | CD95+ CD45ROhi and CD95+ CD45ROlow subsets | Are associated with better prognosis and longer survival in patients with breast tumors. | [51] |
| γδ T cells | Vδ1 and Vγ9Vδ2 | Have anti-tumor properties by differentiating into cytotoxic T cells. | [52-55] |
Tumor-infiltrating T cell subpopulations include CD4+ and CD8+ T cell subsets (Table 2). CD4+ T cells are represented by T helpers, such as TH1, TH2, TH9, TH1 7 and Tfh, and Tregs. The CD8+ T cell subsets consist of cytotoxic cells and mucosal-associated invariant T cells (MAIT). These T cell subpopulations are highly important for prognosis and prediction of treatment efficiency. CD8+ cytotoxic cells migrate into the tumor tissue and display cytotoxic activity against tumor cells [44].
γδ T cells, a T cell subset involved in both innate and adaptive immunity, may have both anti-tumor and pro-tumor functions [56]. Anti-tumor effects are mediated via their differentiation into cytotoxic T cells. Vδ1 and Vγ9Vδ2 T cells are two populations of γδ T cells, which are present in the tumor microenvironment. Both subsets develop cytotoxic capabilities [52]. The lysis of the target tumor cells by γδ T cells is mediated via different mechanisms involving granzyme B, perforin, Fas ligand. In addition, γδ T cells induce cytostatic effects by producing IFN-γ or TNF-α [53, 54]. In contrast to anti-tumor effects, IL-17+ γδ T cells are associated with the progression of ovarian tumors [57]. Based on their functional capabilities, γδ TH1, γδ TH2, γδ TH17, γδ Tfh, and γδ Treg cells can be distinguished among the effector γδ T cells [56]. Notably, these cells can differentiate from one subset to another under the influence of different cytokines.
Among the T helpers, TH9 cells are known to secrete IL-9 and IL-10 and inhibit tumor growth [45]. Low expression of IL-9R induces melanoma growth. Moreover, some TH9 cells can secrete IFNg. Also, TH9 cells, which infiltrate colorectal tumors, may be regulated through PD-1/PD-L1 pathway and may stimulate the proliferation of CD8+ cells. Expansion of TH9 cells is usually accompanied by an increase of IL-9+ IL-4- and IL-9+ IL-4+ cell subsets [58]. Surprisingly, other studies have shown that TH9 cells may stimulate epithelial-mesenchymal transition and dissemination of lung tumor cells [59]. Furthermore, TH9 cells infiltrate lung tumors and are associated with poor survival.
Tregs are known to infiltrate tumor sites and demonstrate immunosuppressive activity. Tumor-infiltrating Tregs were found to have high expression of CTLA-4, GITR and PD-1 [46]. Tregs express multiple chemokine receptors with corresponding ligands, such as CCR with CCL12, CCR4 with CCL17 and CXCR4 with CXCL1 [47]. With regard to phenotypical characteristics, tumor-infiltrating Tregs express immunosuppressive markers, such as iCTLA-4. In addition, these cells have upregulated expression of GITR, LAG3, OX40 and ICOS [48].
Mucosal-associated invariant T cells (MAIT) belong to innate T cells and are present mainly in mucosal tissues. They express a semi-invariant T cell receptor, which contains rearranged TCRβ chains with Vβ gene segments [49]. MAIT cells demonstrate strong cytotoxic activity in colon tumors. The functions of these cells include both production of TH1 -mediated cytokines and direct lysis of neoplastic cells [50]. Circulation of MAIT cells was downregulated in patients with hepatocellular carcinoma and colorectal and lung tumors [60]. Notably, MAIT cells, which infiltrated tumor tissues in patients with colorectal carcinoma, secreted less IFNg than MAIT cells from intact livers.
With regard to memory T cells, Vahidi et al analyzed the presence of CD8+ memory T cell groups in draining lymph nodes from patients with breast tumors. Increased numbers of CD45low central memory T cells were found in lymph nodes, indicating that the early differentiation of memory T cells is inhibited by tumor-produced regulatory factors [61]. The increase of tumor-infiltrating T cells usually correlates with positive outcomes in solid tumors. Tissue-resident memory T cells were associated with better prognosis and longer survival in patients with breast tumors [51].
There are three main subsets of naïve B lymphocytes: follicular B cells, marginal zone B cells and B1 B cells. Mature follicular B cells migrate through blood and lymph, reside in specific B cell areas of lymph nodes, Peyer’s patches, and the spleen and may present T-dependent antigens to T cells. Marginal zone CD19+ CD21+ CD23- CD24+ IgM+ B cells reside in the marginal sinus of the spleen and mediate the transport of antigen in immune complexes. B1 cells are involved in the development of IgM responses to bacterial T cell-independent antigens. These cells can migrate from the peritoneum and reside in mesenteric lymph nodes. Memory B-cells are represented by three subsets: pre-switch IgD+ IgM+ CD27+ B cells, IgD- IgM+ CD27+ B cells, post-switch IgA+ CD27+ and IgG+ CD27+ B cells and IgA+ CD27- and IgG+ CD27- memory B cells [62]. R Shi et al obtained memory B cells specific to SARS-CoV-2 RBD using the following markers: CD3−, CD16−, CD235a−, CD38−, CD19+, CD27+, IgG+ in addition to His+ (for His-tagged SARS-CoV-2 RBD) [63]. Circulating plasmablasts can be identified by the expression of CD38 and CD138 [64].
| Name | Type | Phenotype | Markers to sub-fractionate | Functions |
|---|---|---|---|---|
| Transitional | T1 | IgD+ CD27neg CD10+ CD24high CD38high MTG+ | Precursor to T2; IL10 production (?) | |
| T2 | IgD+ CD27neg CD10+ CD24high/+ CD38high/+ MTG+ | Precursor to T3; IL10 production (?) | ||
| T3 | IgD+ CD27neg CD10neg CD24+/low CD38+/low MTG+ | Precursor to mature-naive; IL10 production (?) | ||
| Mature-naive | IgD+ CD27neg CD10neg CD24+/low CD38+/ low MTGneg | CD23, CD69, CD80, CD86 | Precursor to GC, memory, and antibody-secreting cells | |
| Memory | Double-negative | IgDneg CD27neg | CD21, CD24, CD95, CXCR3 | Recall responses |
| Non-switched | IgD+ CD27+ | CD1c, CD21, CD24 | Immunoprotective self antibody, regulatory | |
| IgM- only | IgM+ IgDneg CD27+ | CD1c, CD21, CD24 | Immunoprotective self antibody, regulatory | |
| Switched | IgMneg IgDneg CD27+ | CD21, CD24, CD95, CXCR3 | Pathogen protection; autoimmune pathology | |
| Antibody-secreting cell | Plasmablast | IgDneg CD27high CD38high CD138neg | CD20, HLA-DR | Antibody secretion |
| Plasma cell | IgDneg CD27high CD38high CD138+ | CD20, HLA-DR | Antibody secretion |
Expression of BCR expression is highly important for maintaining B cells in the peripheral immune system. However, only 30% of B cells in spleen develop into mature B cells. Moreover, mice, which have mutations in genes encoding BCR-related proteins, including BLNK, Btk, and Vav, show disruption of the maturation process [66, 67].
Lymphoid progenitors Lin- KITlow CSA1low IL-7R+ are considered to be a lymphoid progenitor group and can differentiate into both B and T cells. Also, in vitro studies have demonstrated that B220- CD19+ cells can differentiate into myeloid or B cells [68] and Lin- KITlow SCA1low IL-7R+ FLT3+ CD34- cells or B220- KITlow SCA1+ CD24+ CD43+ cells contain increased numbers of B cell precursors [69, 70].
Early B220+ precursors of B cells do not express cell surface immunoglobulin (Ig), reside in the bone marrow and include pre-pro-B cells, pro-B cells, and pre-B cells. Immature pre-B cells migrate to the spleen, where they differentiate into mature B cells and plasmocytes (Figure 3). Peripheral B cell subsets, including transitional, mature, memory and antibody-secreting cells, express different surface markers (Table 3).
In addition to IgG production, a subpopulation of splenic B cells can possess regulatory functions. Regulatory B cells (Bregs) affect various parts of the immune system with IL-10 playing a key role in these processes. The B10 subgroup of B cells was shown to act as regulatory cells in experimental models of lupus and autoimmune encephalomyelitis [71]. Moreover, IL-10 producing Bregs with the surface phenotype CD19+ CD24hi CD38hi were found in the peripheral blood in SLE patients [72]. In addition, regulatory phenotypes CD19+, CD24+ CD27+ and CD19+ IgD+ CD24hi CD38hi CD5hi were shown to have suppressive functions in humans [73, 74].
| B cellular subsets | Markers | Functions | References |
|---|---|---|---|
| Regulatory B cells | CD20+ CD27- PD-L1+ CD19+ CD5+ CD43+ | Suppress CD8+ cytotoxic T cells by producing IL-10. | [75-77] |
| Memory B cells | IgM+ IgD- CD27+ | Support anti-tumor immune response, their presence correlates with better survival. | [78] |
| Activated B cells | CD19+ CD80+ CD86+ CD44+ CD69+ PD-L1+ | Stimulate differentiation of CD4+ T cells into TH1 7 cells via the secretion of IL-27 and IL-6 | [79] |
| Plasma cells | CD20- CD24- CD27hi CD38hi | Produce antibodies, their presence correlates with better survival | [78] |
Several subsets of Bregs were characterized in human peripheral blood. These subsets include B cells with different levels of maturity: transitional CD19+ CD24hi CD38hi Bregs [80, 81], CD19+ CD27int CD38+, plasmablasts [82] and CD19+ CD25+ CD71+ B regulatory 1 cells [83]. Recent studies suggest that differentiation and stimulation of Bregs are likely to be induced by inflammation associated with either infection or autoimmune reactions. In particular, toll-like receptor agonists of bacterial origin were shown to activate Bregs in vitro [84, 85]. In addition, the proliferation of Bregs was reported in a murine model of autoimmune arthritis [86].
In addition to the subsets of Bregs mentioned above, Tim-1+ B cells were also shown to regulate immune reactions, since Tim-1 mucin domain-mutated mice develop autoimmune disorders [87]. Tim-1+ Bregs were identified within different B cell subpopulations, including CD19+ CD1dhi CD5+, MZ and B1 cells [88]. Also, human CD73− CD25+ CD71+ BR1 cells were demonstrated to be involved in the development of allergen tolerance [83]. Membrane regulatory molecules expressed by Bregs include CD25, CD71 and CD274 [72, 89, 90].
| Protein | Top three suppliers | Reference |
|---|---|---|
| B220 | BioLegend 103202 (155), Invitrogen 14-0452-86 (140), BD Biosciences 560777 (69) | [91, 92] |
| c-kit | BioLegend 313201 (19), Invitrogen 14-1172-85 (19), Cell Signaling Technology 3074 (18) | |
| CD1C | BioLegend 331501 (20), Miltenyi Biotec 130-090-695 (9), Abcam ab270797 (1) | |
| CD1D | BD Biosciences 339186 (94), Leica Biosystems PA0554 (14), Ventana 790-4341 (7) | |
| CD3E | Invitrogen MA1-90582 (294), BD Biosciences 339186 (94), BioLegend 300402 (45) | [91, 93] |
| CD4 | Invitrogen MHCD0400 (122), BD Biosciences 555344 (110), BioLegend 300502 (47) | [93] |
| CD5 | Invitrogen MA5-13308 (21), Beckman Coulter IM2637U (7), Leica Biosystems NCL-CD5-4C7 (7) | [93] |
| CD8 | Invitrogen MHCD0800 (154), Dako M7103 (85), BD Biosciences 339188 (85) | [93] |
| CD10 | Invitrogen MA5-14050 (56), Leica Biosystems PA0271 (22), BD Biosciences 555373 (15) | |
| CD11a | Invitrogen MA1-19003 (7), BioLegend 301202 (5), BD Biosciences 555381 (5) | |
| CD19 | BioLegend 302202 (77), BD Biosciences 564457 (67), Invitrogen 14-0199-82 (26) | [91, 94] |
| CD21 | Invitrogen MA5-11417 (11), BD Biosciences 555421 (10), Dako M0784 (6) | |
| CD23 | Invitrogen MA5-14572 (11), Leica Biosystems NCL-CD23-1B12 (4), BD Biosciences 550386 (3) | |
| CD24 | BD Biosciences 555428 (29), Invitrogen MA1-10154 (19), BioLegend 311102 (7) | |
| CD25 | BD Biosciences 560356 (65), BioLegend 302602 (42), Invitrogen 14-0259-82 (18) | |
| CD27 | BioLegend 302839 (36), BD Biosciences 561408 (36), Invitrogen 14-0271-82 (32) | |
| CD28 | BioLegend 302902 (23), BD Biosciences 556620 (20), Invitrogen 16-0289-81 (16) | |
| CD38 | BioLegend 303502 (36), BD Biosciences 564498 (33), Invitrogen MA1-19316 (27) | |
| CD44 | BioLegend 103002 (168), Invitrogen 14-0441-81 (130), Cell Signaling Technology 3570 (36) | |
| CD45RB | BioLegend 103202 (155), Invitrogen 14-0452-86 (140), BD Biosciences 560777 (69) | [95] |
| CD49d | BD Biosciences 555502 (11), BioLegend 304302 (8), Invitrogen 12-0499-42 (6) | |
| CD58 | BD Biosciences 555921 (3), BioLegend 330902 (2), Beckman Coulter IM3702 (2) | |
| CD62L | Invitrogen MA1-10259 (27), BD Biosciences 555542 (25), BioLegend 304802 (17) | |
| CD69 | BioLegend 310902 (35), BD Biosciences 560740 (33), Invitrogen MA1-207 (29) | |
| CD71 | Invitrogen 13-6800 (454), BD Biosciences 555534 (18), BioLegend 334102 (10) | |
| CD73 | BD Biosciences 550257 (51), Santa Cruz Biotechnology sc-25603 (28), BioLegend 344002 (4) | |
| CD80 | BioLegend 305201 (26), BD Biosciences 557223 (26), Invitrogen MA1-19215 (15) | |
| CD86 | BioLegend 305402 (36), Invitrogen MA1-10293 (33), BD Biosciences 555656 (23) | |
| CD95 | BD Biosciences 555670 (27), BioLegend 305602 (14), Invitrogen MA1-20163 (10) | |
| CD99 | Abcam ab22506 (19), Dako M3601 (17), Invitrogen MA5-12287 (5) | |
| CD103 | BioLegend 350202 (9), BD Biosciences 550258 (8), Invitrogen 14-1038-82 (7) | |
| CD130 | Santa Cruz Biotechnology sc-376280 (2), R&D Systems MAB628-100 (1), BioLegend 362006 (1) | |
| CD134 | BD Biosciences 555838 (4), BioLegend 350002 (3), Invitrogen 11-1347-42 (1) | |
| CD138 | Dako M7228 (17), Abcam ab34164 (16), BD Biosciences 650660 (12) | |
| CD161 | BioLegend 339902 (18), BD Biosciences 556079 (11), Miltenyi Biotec 130-092-676 (8) | |
| CD127 | Invitrogen 14-1278-82 (31), BioLegend 351302 (29), BD Biosciences 552853 (25) | |
| CD274 | Cell Signaling Technology 13684 (109), Sino Biological 10084-R015 (36), BioLegend 329701 (21) | |
| CD357 | Invitrogen 14-5875-80 (4), BioLegend 311603 (2), MilliporeSigma SAB1404625 (1) | |
| CCR4 | BD Biosciences 551121 (12), BioLegend 359402 (7) | |
| CCR5 | BD Biosciences 555991 (12), BioLegend 313707 (5), Invitrogen 12-1957-42 (1) | |
| CCR6 | BD Biosciences 559560 (18), BioLegend 353402 (17), Invitrogen 14-1969-82 (4) | |
| CCR7 | BioLegend 353202 (34), BD Biosciences 552174 (34), Invitrogen 14-1979-82 (17) | [96] |
| CCR9 | BD Biosciences 561607 (2) | |
| CCR10 | BD Biosciences 561607 (2) | |
| CLA | BioLegend 321302 (9), BD Biosciences 550407 (4), Invitrogen 17-1629-42 (1) | |
| CXCR3 | BioLegend 353702 (19), BD Biosciences 557183 (9), Invitrogen 12-1839-42 (1) | |
| CXCR4 | Invitrogen 14-9991-82 (19), BioLegend 306502 (16), BD Biosciences 555974 (14) | |
| CRTH2 | BioLegend 350102 (7), BD Biosciences 558412 (7), Beckman Coulter A07413 (1) | |
| FoxP3 | Invitrogen 14-4776-82 (59), Abcam ab20034 (48), BioLegend 320102 (20) | |
| Granzyme A | BioLegend 507202 (9), Abcam ab10870 (1), BD Biosciences 557449 (1) | |
| Granzyme B | Invitrogen MA1-80734 (58), BioLegend 515406 (30), BD Biosciences 561151 (30) | |
| IL-18Ra | BioLegend 313804 (4), Invitrogen 12-7183-42 (1) | |
| KLRG-1 | BioLegend 138429 (7), Invitrogen A14743 (2), Santa Cruz Biotechnology sc-32755 L (1) | |
| PD-1 | BioLegend 329902 (63), BD Biosciences 562138 (18), Invitrogen 14-2799-80 (16) | |
| Perforin | BioLegend 308102 (16), Invitrogen 14-9994-82 (9), BD Biosciences 556434 (8) | |
| SCA1 | Cell Signaling Technology 9664 (586), BD Biosciences 559565 (47), Novus Biologicals NB100-56708 (42) | |
| TCF7 | Cell Signaling Technology 2203 (34), BD Biosciences 564217 (3), Santa Cruz Biotechnology sc-101170 (2) | [96] |
Activated B cells, regulatory B cells and memory B cells are among the most notable B cell subpopulations found within tumor tissues. The functions of tumor-infiltrating heterogenous B lymphocyte subsets include antigen presentation, secretion of antibodies and cytokines, and activation of T cells. In particular, B cells can promote T cell proliferation by functioning as antigen-presenting cells [97]. Tumor-specific antibodies, which recognize tumor antigens, may potentially be used to reduce tumor cells or activate cytotoxic T cells [98]. However, regulatory B cells may act as immune suppressors thus playing a protumor role in the tumor loci. These negative functions of regulatory B cells led to the promotion of therapeutic strategies based on B cell depletion [99].
The detection of B-lymphocytes in the tumor microenvironment has been associated with better prognosis in several tumors (Table 4). In particular, the increased numbers of CD20+ cells were associated with better survival in patients with pancreatic ductal adenocarcinoma [100]. A recent study analyzed different subsets of B cells present in tumor micronvironment [101]. Increased numbers of tumor-infiltrating naive B cells (CD20+ CD27- IgM+ ), IgM+ memory B cells (CD20+ CD27+ IgM+ ), CD27- isotype-switched memory B cells (CD20+ CD27- IgM- ), and plasma cells (CD20- CD24- CD27hi CD38hi ) were found to be associated with better survival in hepatocellular carcinoma [78].
With regard to regulatory subsets, Wu et al found elevated numbers of CD20+ CD27- PD-L1+ regulatory B cells, which were positively associated with melanoma stage [75]. These PD-L1+ cells were suggested to act as T cell inhibitors. Furthermore, regulatory B cells suppress CD8+ cytotoxic activity by producing IL-10 [76]. In line with these data, CD19+ CD5+ CD43+ B1a Bregs were shown to suppress anti-melanoma immune response by secreting IL-10 and inhibiting TH1 cytokine release by cytotoxic CD8+ T cells [77]. The selective inhibition of mitogen-activated protein kinase (MAPK) kinase (MEK), a crucial member of RAS pathway, suppresses regulatory B cells in lymph nodes without affecting humoral immunity [102].
In addition, IgD- CD27+ memory, CD86+ CD21- antigen-presenting and CD86+ activated B cells also infiltrated tumor tissues. Activated naïve B cells, which expressed elevated levels of CD80, CD86, CD44, CD69, and PD-L1, were found to suppress TH1 7-cell expansion through the PD-1/PD-L1 pathway and stimulated differentiation of CD4+ T cells into TH1 7 cells via the secretion of IL-27 and IL-6 [79]. Also, in a study of B cell subsets in hepatocellular carcinoma, the authors have shown that increased numbers infiltrating CD20+ B cells, IgM+ memory B cells, CD27- isotype-switched memory B cells and plasma cells correlated with better survival [78].
One of the most important functions of B cells is antibody production. Enzyme-linked immunosorbent assay (ELISA) can analyze secreted antibodies, plaque-forming cell (PFC) assays can detect antibody-secreting B cells, and ELISPOT can indicate the number of antibody-producing B cells.
Labome surveys formal publications to develop Validated Antibody Database (VAD). Table 5 lists the most cited antibodies against T cell markers and B cell markers among the 60,000 articles Labome has surveyed.
CD45, also called leukocyte common antigen(LCA), regarded as a pan-immune marker, has also been found in rare epithelial cells in mouse intestine [103], more specifically in tuft-2 cells [104].
- Germain R. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309-22 pubmed
- Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317-30 pubmed
- Wang R, Xie H, Huang Z, Ma J, Fang X, Ding Y, et al. Transcription factor network regulating CD(+)CD8(+) thymocyte survival. Crit Rev Immunol. 2011;31:447-58 pubmed
- Pepper M, Jenkins M. Origins of CD4(+) effector and central memory T cells. Nat Immunol. 2011;12:467-71 pubmed
- Asarnow D, Kuziel W, Bonyhadi M, Tigelaar R, Tucker P, Allison J. Limited diversity of gamma delta antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell. 1988;55:837-47 pubmed
- Kyes S, Carew E, Carding S, Janeway C, Hayday A. Diversity in T-cell receptor gamma gene usage in intestinal epithelium. Proc Natl Acad Sci U S A. 1989;86:5527-31 pubmed
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057-61 pubmed
- Chen W, Jin W, Hardegen N, Lei K, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875-86 pubmed
- Davidson T, DiPaolo R, Andersson J, Shevach E. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022-6 pubmed
- Levings M, Sangregorio R, Roncarolo M. Human cd25(+)cd4(+) t regulatory cells suppress naive and memory T cell proliferation and can be expanded in vitro without loss of function. J Exp Med. 2001;193:1295-302 pubmed
- Lehmann J, Huehn J, de la Rosa M, Maszyna F, Kretschmer U, Krenn V, et al. Expression of the integrin alpha Ebeta 7 identifies unique subsets of CD25+ as well as CD25- regulatory T cells. Proc Natl Acad Sci U S A. 2002;99:13031-6 pubmed
- Li Z, Mahesh S, Kim B, Buggage R, Nussenblatt R. Expression of glucocorticoid induced TNF receptor family related protein (GITR) on peripheral T cells from normal human donors and patients with non-infectious uveitis. J Autoimmun. 2003;21:83-92 pubmed
- Solis Castillo L, García Romo G, Díaz Rodríguez Á, Reyes Hernández D, Tellez Rivera E, Rosales Garcia V, et al. Tumor-infiltrating regulatory T cells, CD8/Treg ratio, and cancer stem cells are correlated with lymph node metastasis in patients with early breast cancer. Breast Cancer. 2020;: pubmed publisher
- Kurosaki T. Functional dissection of BCR signaling pathways. Curr Opin Immunol. 2000;12:276-81 pubmed
- Kurosaki T. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol. 1999;17:555-92 pubmed
- Montecino Rodriguez E, Leathers H, Dorshkind K. Bipotential B-macrophage progenitors are present in adult bone marrow. Nat Immunol. 2001;2:83-8 pubmed
- Tudor K, Payne K, Yamashita Y, Kincade P. Functional assessment of precursors from murine bone marrow suggests a sequence of early B lineage differentiation events. Immunity. 2000;12:335-45 pubmed
- Mojica M, Perry S, Searles A, Elenitoba Johnson K, Pierce L, Wiesmann A, et al. Phenotypic distinction and functional characterization of pro-B cells in adult mouse bone marrow. J Immunol. 2001;166:3042-51 pubmed
- Evans J, Chavez Rueda K, Eddaoudi A, Meyer Bahlburg A, Rawlings D, Ehrenstein M, et al. Novel suppressive function of transitional 2 B cells in experimental arthritis. J Immunol. 2007;178:7868-78 pubmed
- Materials and Methods [ISSN : 2329-5139] is a unique online journal with regularly updated review articles on laboratory materials and methods. If you are interested in contributing a manuscript or suggesting a topic, please leave us feedback.
- methodgene
- human ACKR2
- human C-kit
- human CCR4
- human CCR5
- human CCR6
- human CCR7
- human CD10
- human CD19
- human CD1C
- human CD1D
- human CD21
- human CD23
- human CD24
- human CD25
- human CD27
- human CD274
- human CD28
- human CD38
- human CD3E
- human CD4
- human CD44
- human CD45
- human CD5
- human CD58
- human CD62L
- human CD69
- human CD73
- human CD80
- human CD86
- human CD8A
- human CD99
- human CRTH2
- human CXCR3
- human CXCR4
- human FOXP3
- human Fas
- human GITR
- human GZMA
- human IL18R1
- human IL2RB
- human IL2RG
- human IL7R
- human ITGA4
- human ITGAE
- human KLRB1
- human KLRG1
- human LFA-1
- human OX40
- human PDCD1
- human PRSS3
- human SELPLG
- human TCF7
- human caspase-3
- human gp130
- human granzyme B
- human perforin
- human syndecan
- human transferrin receptor
