This is a knockout-validated antibody summary, based on the publications "Cancer-Specific Loss of p53 Leads to a Modulation of Myeloid and T Cell Responses" for western blot knockout validation (figure s1a) [6], "Stabilization of p21 by mTORC1/4E-BP1 predicts clinical outcome of head and neck cancers" (western blot knockout validation, see figure 1C in the article) [7], The p53R172H mutant does not enhance hepatocellular carcinoma development and progression" [1], "The MDM2-p53-pyruvate carboxylase signalling axis couples mitochondrial metabolism to glucose-stimulated insulin secretion in pancreatic β-cells" [2], "The tumour suppressor CYLD regulates the p53 DNA damage response" [3], "Tracking and transforming neocortical progenitors by CRISPR/Cas9 gene targeting and piggyBac transposase lineage labeling" [8], "Creation and preliminary characterization of a Tp53 knockout rat" [4], "An inducible long noncoding RNA amplifies DNA damage signaling" [9] (see figure 1d in the article), "Targeted Deletion of p53 in the Proximal Tubule Prevents Ischemic Renal Injury" [5], "Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer" for western blot knockout validation (see figure 5h [10] ), "Rapid interrogation of cancer cell of origin through CRISPR editing" for (figure 2a, 5b,5d) [11], and "Tumor genotype dictates radiosensitization after Atm deletion in primary brainstem glioma models" for western blot knockout validation (figure s3) [12]. Labome curates formal publications to compile a list of antibodies with unambiguous specificity within Validated Antibody Database (VAD).
Cell Signaling
Antibody: Mouse monoclonal p53
Catalogue No: 2524
Clone: 1C12
Summary: Mouse monoclonal antibody raised against a synthetic peptide corresponding to residues surrounding Ser20 of human p53.
Supplier recommended for WB, IP, IF, flow cytometry and ChIP.
Reacts with human, mouse, rat, hamster and monkey p53.
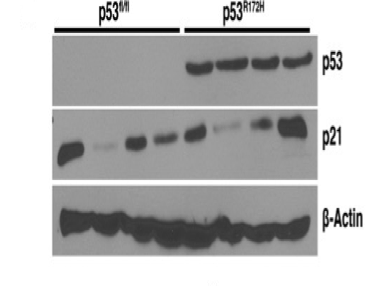
Technique: WB (Figure 1).
Sample: tumor cell line
Dilution :1:2000
This antibody was validated in p53 flox/flox mice by immunoblotting [1]. P53 expression in the cell line derived from of p53 flox/flox mice was not detected by western blot.
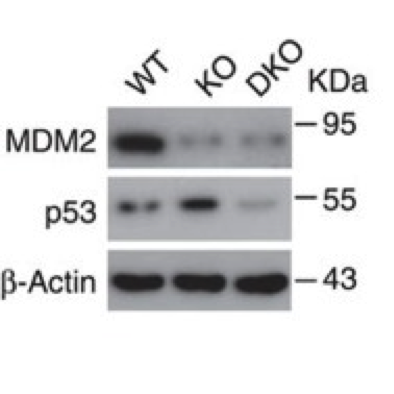
Western blot (Figure 2).
Isolated pancreatic islets from wild-type and knockout mice.
dilution is 1:1000.
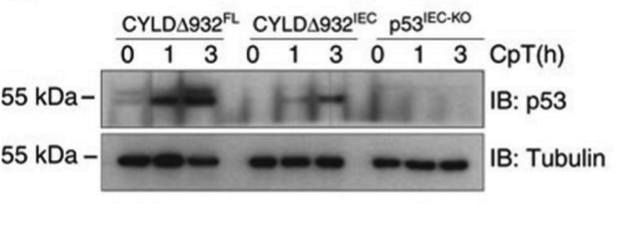
Western blot (Figure 3)
Intestinal epithelial organoids obtained from CYLDΔ932FL, CYLDΔ932IEC, and p53IEC-KO mice.
1:750 dilution at 4° O/N.
Horseradish peroxidase-coupled antibodies (GE Healthcare and Jackson Immune Research).
Chemiluminescent detection substrate (GE Healthcare and Thermo Scientific).
Immunohistochemistry
Control and CRISPR/Cas9 targeting P53 Wistar rats brains. Animals were perfused transcardially with 4% paraformaldehyde/PBS (4% PFA) and brain samples were post-fixed overnight in 4% PFA. Brains were sectioned at 65 µm thickness on a vibratome (Leica VT 1000S) and processed as free-floating sections.
1:50 dilution.
Zeiss Axio imager M2 microscope with Apotome with 488/546/350 nm filter cubes and the X-Cite series 120Q light source.
Please see Suppl. Figure 1f,g in the article [8].
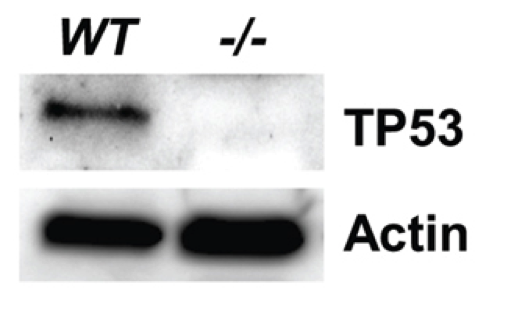
Western blot (Figure 4).
Primary cultured tail cells from both wild-type (WT) and Tp53Δ11/Δ11 (−/−) rats. Cells were lysed in RIPA buffer (Sigma) containing 1× Protease Inhibitor Cocktail (Sigma).
5% milk for 30 minutes.
1:500 dilution in 5% milk solution for 1 hour.
1:10,000 dilution goat anti-mouse (Jackson ImmunoResearch, West Grove, PA) for 30 minutes.
Supersignal West Pico substrate (Thermo Scientific, Waltham, MA) for 2 minutes and visualized on a ChemiDoc XRS+ (Bio-Rad).
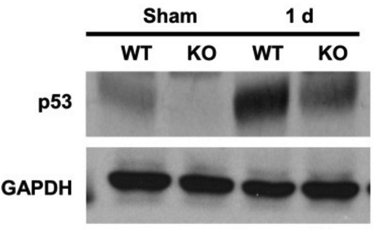
Western blot (Figure 5).
Whole-renal tissue extracts from control and kidney proximal tubule-specific p53-KO mice.
Overnight at 4°C.
Horseradish peroxidase-conjugated secondary antibodies (1:5,000; Vector Laboratories).
Western Lighting Plus-ECL (NEL104001EA; PerkinElmer, Waltham, MA), and x-ray film.
- If you are aware of any publication with knockout studies validating a monoclonal or recombinant antibody, either purchased from a supplier or developed by the author(s), please notify us through feedback.
